|
Recently, scientists discovered by accident that a small strip of Whatman no. 1 paper can capture nucleic acids rapidly from a crude mixture containing ground up tissue in a lysis buffer. The paper strip can then be quickly washed without releasing the nucleic acid. After these steps, the paper strip retains enough nucleic acids to be dipped into a PCR reaction mixture and be used for amplification (Zou et al. 2017). Ultimately they created a dipstick protocol to extract DNA in 30 seconds to use in a PCR amplification reaction. Excellent for low-tech environments and the field! Maybe this can be optimized at some point to be a part of ONT nanopore-sequencing pipelines, to make sequencing indeed available to everyone, at any moment, anywhere. For now, this seems a very cool way to do quick population screening of microbes in soil for example! See below for their dipstick protocol! Zou et al. 2017, Nucleic acid purification from plants, animals and microbes in under 30 seconds, Plos Biology. https://doi.org/10.1371/journal.pbio.2003916 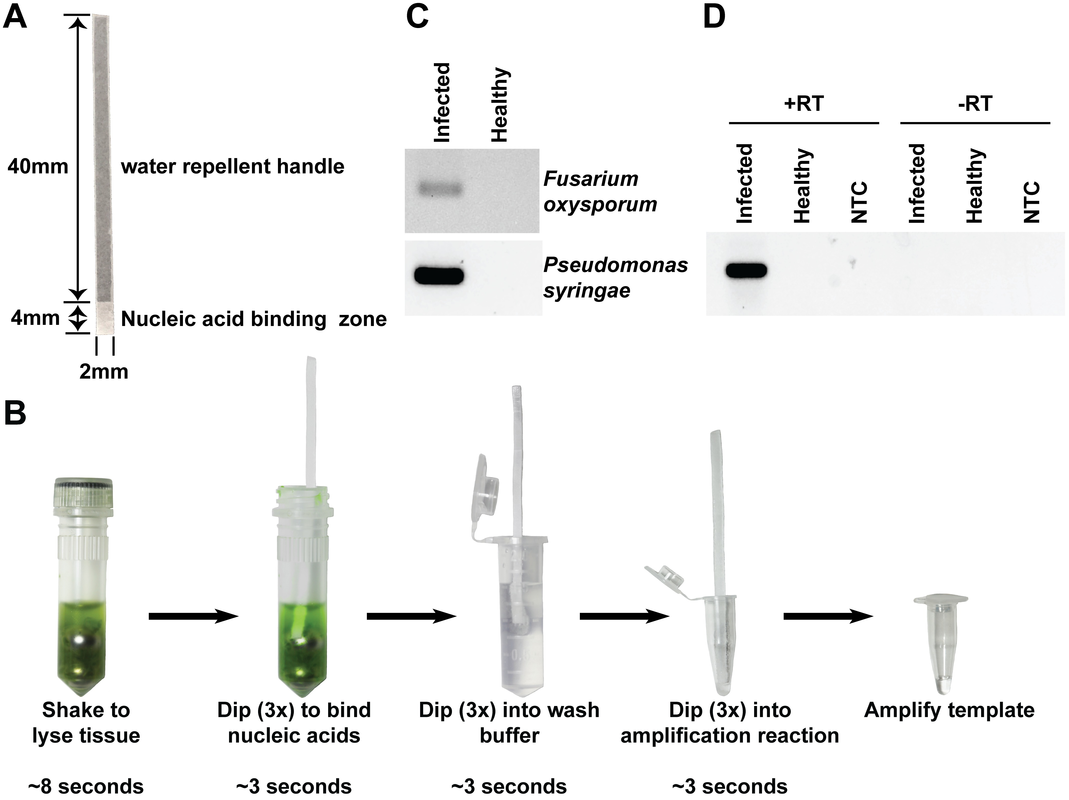 (A) The cellulose dipstick consists of a 2x40 mm wax impregnated handle and a 2x4 mm nucleic acid binding zone. (B) An overview of the dipstick-based purification method in which tissue is homogenised by shaking it in a tube containing ball bearings and an appropriate extraction buffer. The dipstick is used to bind the nucleic acids by dipping it three times into the homogenate, washed by dipping it three times into a wash buffer, and eluted by dipping it three times in the amplification reaction mix. (C) Nucleic acids were purified using the cellulose dipstick method from Arabidopsis leaves infected with F. oxysporum f.sp. conglutinans (upper image) or P. syringae (lower image) and eluted into PCR reaction mixes containing pathogen-specific primers. (D) Nucleic acids were purified from tomato leaves infected with Cucumber mosaic virus using the cellulose dipstick method. The purified DNA was eluted directly into PCR amplification reaction mixes with (+RT) or without (-RT) AMV reverse transcriptase. NTCs involved adding 1 μl of water instead of using dipstick-purified nucleic acids. AMV, Avian myeloblastosis virus; NTC, no template control; RT, reverse transcriptase.
1 Comment
|
Archives
September 2018
Categories
All
|
 RSS Feed
RSS Feed






